What Happens When We Expand the Chronology and Geography of Plague’s History?
(Or Why Yersinia pestis is a Good ‘Model Organism’ in These Pandemic Times)
This short essay is a companion to, and summary of, a lecture of the same title given by Monica H. Green at Oxford on 16 March 2020. You can now watch the video of the lecture on the Oxford Medieval Studies YouTube Channel.
Planning for the following presentation was done in early January 2020, before almost anyone had a clue that the world was about to be immersed in a new pandemic event. Originally meant as a presentation to Byzantinists, the talk was reframed to capture the essence of the larger work I have been doing the past 24 years to reframe the history of infectious diseases in a global framework of analysis. Although still focused on the history of plague (the main disease I’m working on at the moment), my central argument is that an evolutionary approach to the history of infectious diseases gives a powerful new way to understand pandemics past and present, and hopefully a way to avert similar events in the future.
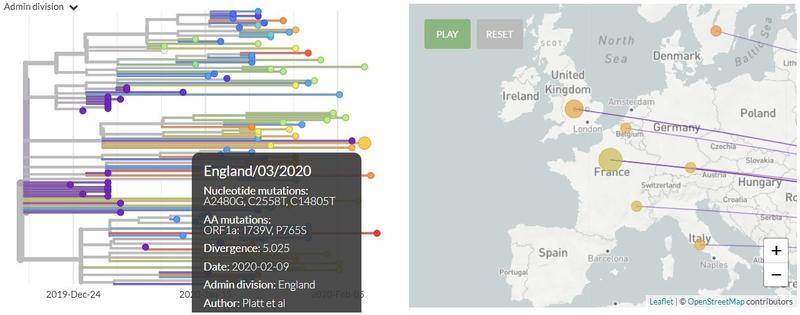
The linchpin of all this work are new ways the biological sciences are contributing to the investigation of disease history. These contributions are of several kinds, but the most important have come from genetics. These function at two levels. First is phylogeny, the work of constructing “family trees” showing the evolutionary development—the “familial” relations—of microorganisms. Everyone will likely have noticed the phylogenetic trees published almost daily for the new coronavirus, SARS-CoV-2. Here’s the one from 9 February, showing genomes just sequenced in England:
SARS-CoV-2 has only been around since about November 2019, so far as we know right now. But prior to its arrival, there have been major diseases circulating around the globe for centuries. Tuberculosis, for example, although almost certainly originating in the eastern hemisphere, was present in the western hemisphere (the Americas) for at least the past 1000 years.
The present talk focuses on the story of plague, the disease caused by the bacterium Yersinia pestis. The talk focuses on summarizing the ways in which genetics—by reconstructing the evolutionary history of the pathogen—has told us things about plague’s history that we never previously had an inkling of. It has told us how deep plague’s history with human populations has been (back to the Late Neolithic). It has suggested how broadly plague may have spread in the past (across the Eurasian steppe in the Bronze Age; throughout east, central, and western Eurasia and even into Africa in Antiquity and the Middle Ages). Here’s the latest phylogenetic tree of Yersinia pestis, with the major pandemic events marked:
Source: Zhemin Zhou, Nabil-Fareed Alikhan, Khaled Mohamed, Yulei Fan, the Agama Study Group, and Mark Achtman, “The EnteroBase use’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity, Genome Research 30 (2020), 138-152, fig. 5. Labeling: M. H. Green, 2020.
Note the shaded areas: the Neolithic transition; the Justinianic Plague; and the Black Death. Historians have known for generations of the latter two events, of course, because we have eye-witness testimony of both events, as well as many other kinds of evidence. What is stunning, however, is that we also now also have aDNA for those events. “aDNA”—the other new kind of evidence genetics has put on the table—is short for “ancient DNA,” molecular fragments that have been retrieved from people who died of the plague. This has been pieced together fragment-by-fragment, allowing us to understand how those strains of Y. pestis compare to modern strains (all the non-shaded circles on the tree). That comparison, in turns, allows us to make inferences about where the different historical strains circulated, and begin to investigate what animal species hosted them.
But telling the story of a one-celled bacterium is only part of the history we need to reconstruct. How was the disease transmitted over such long distances? Why at particular times, but not others? Getting the “human” part of these stories connected to the history of the pathogen is the work that historians now need to do. The present talk, therefore, gives a sketch of what we know about plague’s history and what questions are currently being investigated. In the question-and-answer session at the end, we covered a variety of topics, some having to do with the specifics of plague’s history (especially the many questions we still have about the Justinianic Plague of Antiquity) but also the urgent questions we face in the present day, faced with a pandemic event that gives every sign of being as cataclysmic for its implications on world history as the Black Death of the later Middle Ages.
Humankind has faced pandemics before. We don’t know nearly as much as we should about them, however. There is much to do in making better sense of pandemic events of the past and in better understanding what they might teach us to better face an uncertain future.
Sources for more information:
I’ve written up a teaching guide about the “new genetics paradigm” which explains in more detail what has happened in genetics in the past couple of decades and why it has been so transformational for our “pandemic thinking.” This and information on my other publications on plague can be found at the following link: https://independentscholar.academia.edu/MonicaHGreen/Plague-Studies.
Monica H. Green is a historian of medicine and health. Follow her on Twitter @monicaMedHist and get in touch at mhgreen@asu.edu.
For more medieval matters from Oxford, have a look at the website of the Oxford Medieval Studies TORCH Programme and the OMS blog!